 RNA-KG v2.0 is an ontology-based knowledge graph which encompasses biological knowledge about RNA molecules gathered from more than 90 linked open data sources and ontologies,
integrating functional relationships with genes, proteins, and chemicals and ontologically grounded biomedical concepts.
Relationships are characterized by standardized properties that capture the specific context (e.g., cell line, tissue, pathological state) in which they have been identified.
In addition, the nodes are enriched with detailed attributes, such as descriptions, synonyms, and molecular sequences sourced from platforms such as OBO ontologies, NCBI repositories, RNAcentral, and Ensembl.
RNA-KG is constantly maintained and updated with new experimental data and is a core project of the Monarch Initiative KG-Hub and an RNAcentral expert database.
More details can be found in our pre-print.
RNA-KG v2.0 is an ontology-based knowledge graph which encompasses biological knowledge about RNA molecules gathered from more than 90 linked open data sources and ontologies,
integrating functional relationships with genes, proteins, and chemicals and ontologically grounded biomedical concepts.
Relationships are characterized by standardized properties that capture the specific context (e.g., cell line, tissue, pathological state) in which they have been identified.
In addition, the nodes are enriched with detailed attributes, such as descriptions, synonyms, and molecular sequences sourced from platforms such as OBO ontologies, NCBI repositories, RNAcentral, and Ensembl.
RNA-KG is constantly maintained and updated with new experimental data and is a core project of the Monarch Initiative KG-Hub and an RNAcentral expert database.
More details can be found in our pre-print.
⚠️ Disclaimer: This is the RNA-KG version 2.0 project page.
For the previous release, see
https://RNA-KG.anacleto.di.unimi.it.
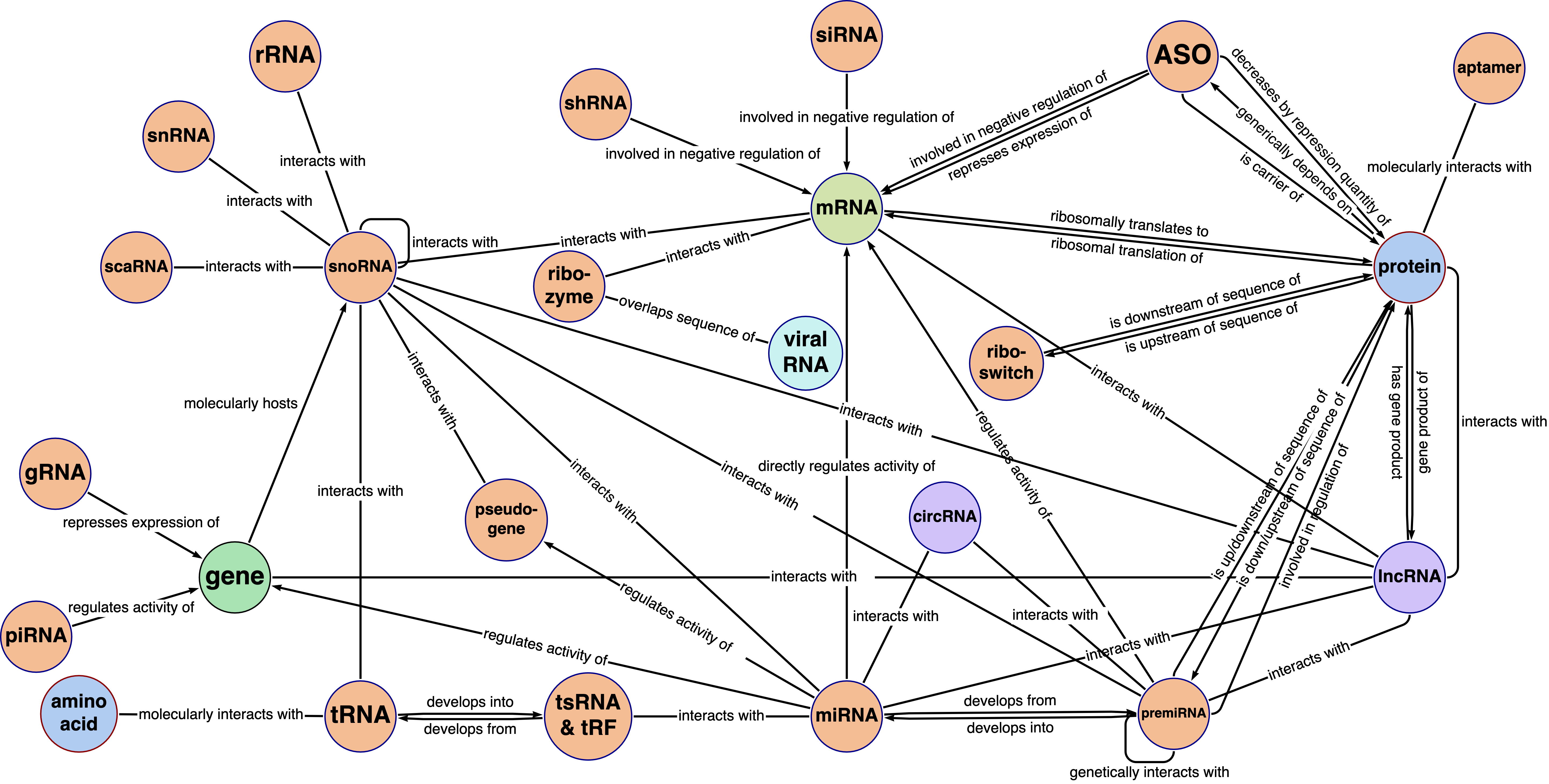
Excerpt of the RNA-KG schema
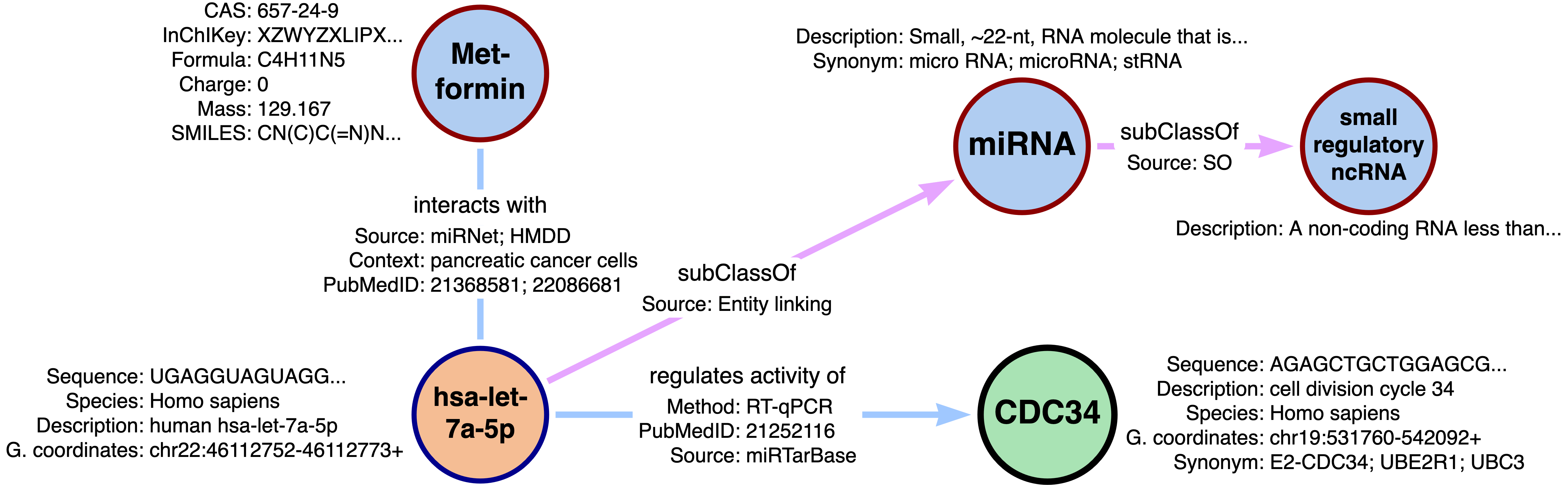
Excerpt of a RNA-KG subgraph
Useful Links
- Public Neo4j endpoint (usr: rnakgv20; pwd: rnakgv20). The database dump is available at: rnakgv20.dump; the list of RNA-KG nodes including properties is stored in: nodes.csv; the list of RNA-KG edges including properties is stored in: edges.csv.
- GitHub repository: collection of tutorials to build RNA-KG v2.0 from scratch (specifically, notebooks contains Python notebooks to build RNA-KG v2.0) and for reproducing experiments such as "context-aware" KG pruning, clustering, and link predictions. Data for reproducing RNA-KG contruction and experiments are available on Zenodo.
- Views already generated by our team. These views are thought to be used in combination with graph-oriented ML techniques for edge and node type labeling and heterogeneous/homogeneous link predictions tasks. For each view, we provide the correspondent schema (in xlsx) and two csv files specifying nodes and edges to facilitate the import in graph-oriented ML libraries such as GRAPE.
Please cite the following papers if RNA-KG was useful for your research:
